
How to use this server:
| Step1: Upload your PDB file or input a PDB ID. The file may contain multiple structures (such as an NMR ensemble or a mulitmeric complex). If you wish to select a particular chain, please include a chain identifier in the PDB ID or edit the PDB file so that it only contains a single chain. Note that ResProx does not calculate or estimate the resolution of DNA or RNA molecules. |
 |
| Step2: Once you have entered the PDB file, press the "Submit" button. The following file loading page sg page. |
 |
| Step3: Once the file loading has been completed a processing "time" bar will appear as shown below. |
 |
| Step4: The results will appear in 10-15 seconds (on an average run). This will take longer if multiple structures are run. Note that ResProx generates an estimated resolution for each model structure. Shown below you see two resolution values. An average and a standard deviation is calculated as well. This is particularly useful when evaluating NMR ensembles or multimeric complexes from X-ray studies as it allows one to identify "good" and "bad" models or chains on an individual basis. |
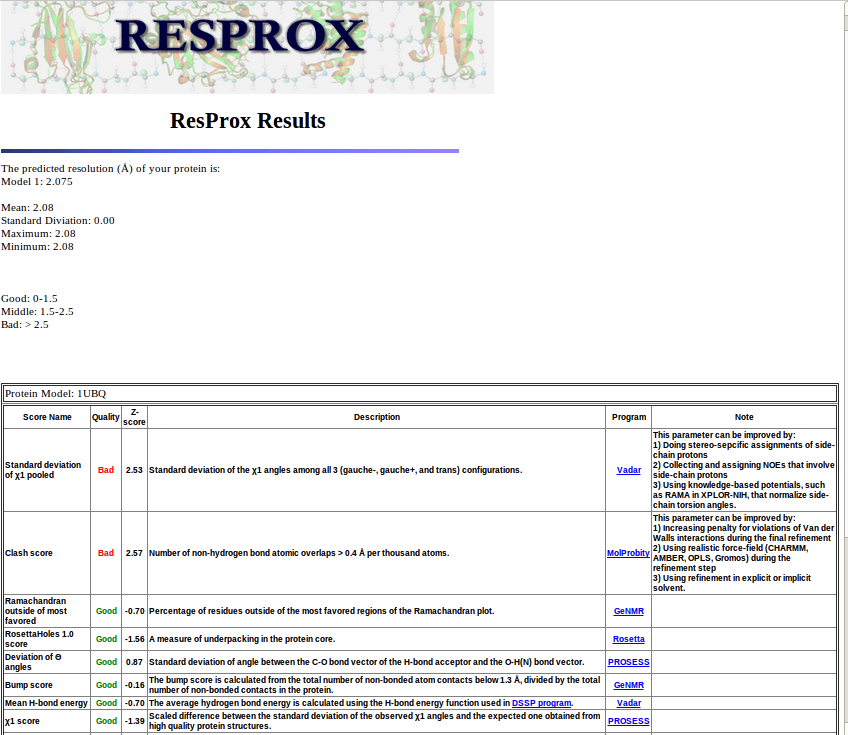 |
How to use URLAPI:
| Step1:Integrate this URLAPI into your program: http://www.resprox.ca/cgi-bin/resprox_command_line.cgi?pdbid=<PDBID>&cid=<CHAIN>(The '<' and '>' is not needed) |
Step2:The reply from the above URLAPI will be like thus:
Your case will finish in 30 seconds up to 5 minutes depending on the size of the PDB file.
Please check this link later for the result. http://www.resprox.ca/cgi-bin/Results.cgi?file=/apps/resprox/tmp/<case_number>/<case_number>(The '<' and '>' is not needed) |
| Step3:User can download or review the result with the above link |
| Note:This URLAPI does not support file submission. If want to submit files, please use the front end of the server. |
|


